Statnet: The Fastest Way to Create a Network
tl;dr:
- Create vertex attributes first, then add in edges (“Piecewise by Node” below)
- The “standard” way is actually the slowest way
- Do not use
network[as.matrix(edgelist)] <- 1
Details
I’ve started working in statnet again, for a project at
work. Because I can’t load the entire social network into memory, I’ve
been doing random samples of (very small) sub-networks. There are
several ways to create networks in statnet, and I wanted to find the
fastest way.
For the timing work, I used a single network of about 10K vertices, with a single vertex attribute, and about 750K edges. I ran this on a fairly powerful machines: 24 cores, 128GB of RAM, 1TB SSD, running CentOS Linux 7.4. I used R 3.4, and SNA version 2.4. All of the computation around creating the network, numbering nodes and so forth was done prior to timing.
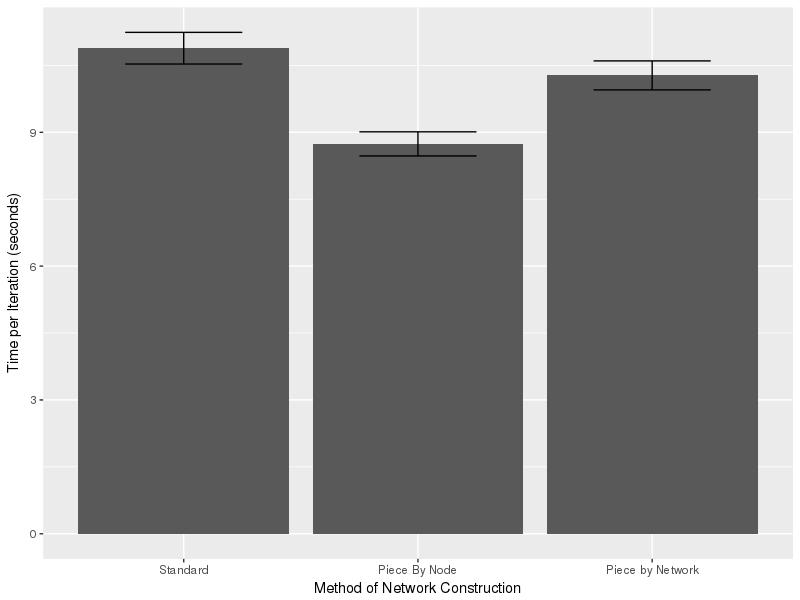
I looked at five ways to create a network in statnet:
- “Standard:” Use the standard statnet constructor and add in a vertex attribute
net <- network(edgelist)
set.vertex.attribute(net, "gender", id_list$gender)
- “Piecewise By Node:” Create a network with nodes, then add in attributes and edges
net <- network.initialize(num_nodes)
set.vertex.attribute(net, "gender", id_list$gender)
network.edgelist(edgelist, the_net)
- “Piecewise By Node (R-Style):” Like “Piece by Node,” but a different syntax for setting the edgelist
net <- network.initialize(num_nodes)
set.vertex.attribute(net, "gender", id_list$gender)
net[as.matrix(edgelist)] <- 1
- “Piecewise By Network:” Create a network, add edges and finally add attributes
net <- network.initialize(num_nodes)
network.edgelist(edgelist, the_net)
set.vertex.attribute(net, "gender", id_list$gender)
- “Piecewise By Network (R-Style):” Like #4, but using the very R-like syntax
net <- network.initialize(num_nodes)
net[as.matrix(edgelist)] <- 1
set.vertex.attribute(net, "gender", id_list$gender)
For each way, I ran it against the same network 50 times to smooth out
any differences, taking care to delete the network (net <- NA) and
run the garbage collector (gc()) between iterations.
Results
First, I didn’t even bother running the R-Style assignments. I let Method #3 run for over 2 hours, and it didn’t even finish the first iteration. After that experience, I didn’t bother with Method #5. That leaves just “Standard,” “Piece by Node” and “Piece by Network” to examine. There’s a pretty big range in times; the slowest run is nearly 50% slower than the fastest run.
There was a clear winner: Piecewise by node was faster than any of the alternates. It was about 15% faster than the next fastest, and 20% faster than the slowest (not counting the horrible “R-style”). The results are shown below with 95% CI.